Pan Wang and al.
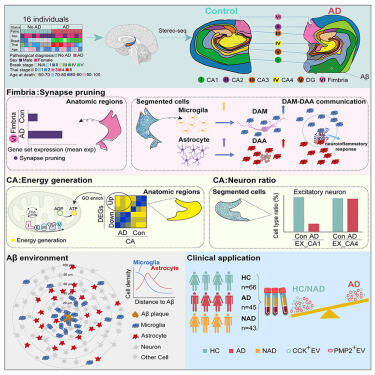
The authors employed the spatial transcriptomics technique Stereo-seq combined with scRNA-seq to investigate the gene expression and cell composition changes in human hippocampus with or without Alzheimer’s disease (AD). The transcriptomic map, with single-cell precision, unveiled AD-associated alterations with spatial specificity, which include the following: (1) elevated synapse pruning gene expression in the fimbria of AD, with disrupted microglia-astrocyte communication likely leading to disorganized synaptic structure; (2) a globally increased energy generation in the cornu ammonis (CA) region, with varying degrees across its subregions; (3) a significant reduction in the number of CA1 neurons in AD, while CA4 neurons remained largely unaffected, potentially due to gene alterations in CA4 conferring resilience to AD; and (4) aggravated amyloid-beta (Aβ) plaques in CA1 and stratum lucidum, radiatum, and moleculare (SLRM). Integration of Stereo-seq map with Aβ staining revealed a sequential enrichment of microglia and astrocytes around Aβ plaques. Finally, reduced brain-derived extracellular vesicles carrying cholecystokinin (CCK) and peripheral myelin protein 2 (PMP2) in AD plasma highlighted their diagnostic potential for clinical applications.